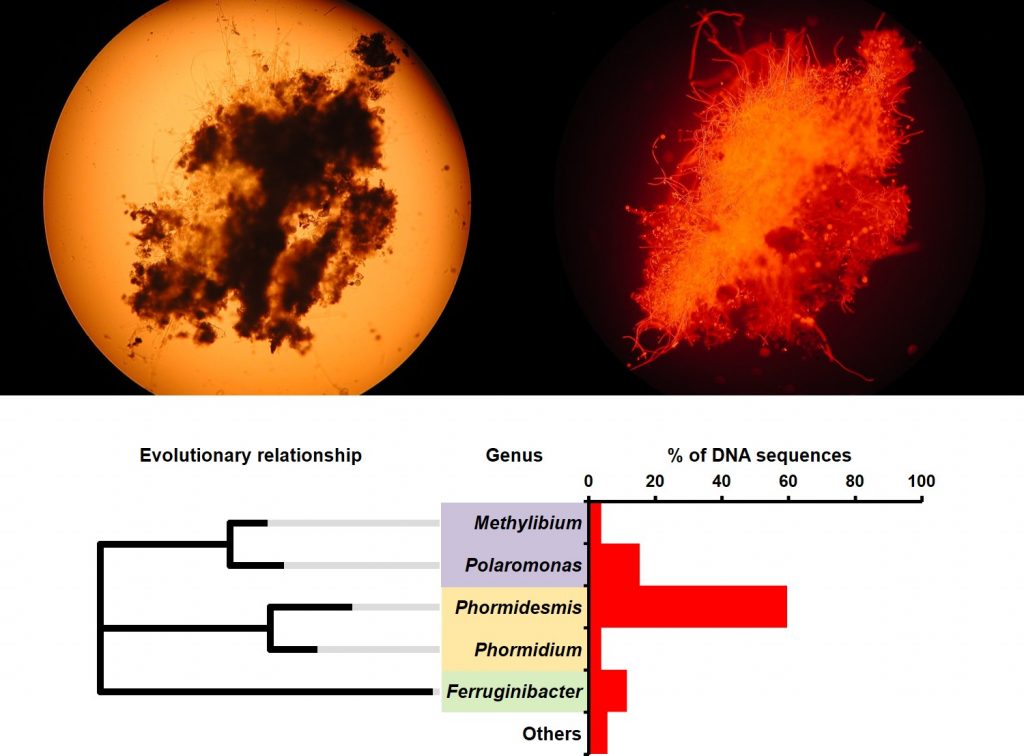
Fig 1: A single grain of cryoconite (top left) is home to a microscopic city of microbes, revealed here by chlorophyll fluorescence microscopy – a technique that causes photosynthesising microbes to emit light (top right) and portable DNA sequencing (bottom panel) [credit: Arwyn Edwards]
Microbes growing on glaciers are recognized for their importance in accelerating glacier melting by darkening their surface and for maintaining biogeochemical cycles in Earth’s largest freshwater ecosystem. However, the microbial biodiversity of glaciers remains mysterious. Today, new DNA sequencing techniques are helping to reveal glaciers as icy hotspots of biodiversity.
To see a world in a grain of…cryoconite
Earth’s glaciers and ice sheets are among its most impressive features, yet this majesty conceals their microscopic riches. We must turn to the microscope and the DNA sequencer to reveal the natural history of glaciers. Rather than a grain of sand, this world lies hidden in a grain of cryoconite. Cryoconite ecosystems are microbe-mineral aggregates which darken the surface of glaciers world-wide which – along with algae – enhance absorption of solar energy and promote glacier melting through so-called bioalbedo feedbacks. Microscopy studies from the late 19th and early 20th century reveal that a diverse range of algae, cyanobacteria, heterotrophic bacteria, protists, fungi and even tardigrades live within cryoconite, but it is only in the last decade that we have started to resolve the genetic diversity of life within cryoconite.
From glaciers to genomes…and back again
Considering glaciers are Earth’s largest freshwater ecosystems, we know very little about the genetic diversity of their inhabitants. Of all known glaciers, fewer than 0.05% have any form of DNA datasets associated with them. Such DNA datasets are commonplace for other environments, as demonstrated by the Earth Microbiome Project. From the limited studies performed, it appears the microbial ecosystems of glaciers are no less diverse than temperate environments: even dark, cold and isolated subglacial lakes harbour thousands of bacterial species. As climatic warming increasingly threatens glaciers, unpicking the interactions between microbes and melt is vital, as is establishing the extent to which glacier biodiversity is threatened. Sequencing microbial genomes from glacial ecosystems is therefore urgent.

Fig 2: Preserving microbial samples from cryoconite for return to the author’s home lab is a conventional approach to studying genetic diversity on glaciers, but the portability of MinION DNA sequencing brings the lab to the field [Credit: Arwyn Edwards].
A DNA sequencer in your rucksack
Such genetic studies have required the collection of samples from glaciers and their return to state-of-the-art laboratories equipped with high throughput DNA sequencers. However, new, portable DNA sequencers are being trialled on cryoconite to permit sequencing of DNA in field labs. Using a pocket-sized DNA sequencer called a MinION connected to the USB port of a laptop, it is possible to extract, sequence and analyse microbial genomes while still in the field. While Nanopore DNA sequencing using MinION devices are increasingly applied to medical emergencies such as Ebola or antibiotic resistance, their highly portable nature means that glacier scientists will be able to collect and analyse microbial genomes while in the field, making the genetic diversity of glaciers accessible.
Who’s who in cryoconite?
Using MinION for DNA sequencing in a field lab at the UK Arctic Station in Ny Ålesund, it was possible to generate rapid profiles of microbial diversity in cryoconite. So who lives in Arctic cryoconite? The most abundant bacterial group identified is a close match to Phormidesmis priestleyi, a filamentous cyanobacterium responsible for engineering the growth of cryoconite grains on Arctic glaciers. In Figure 1 above, Phormidesmis is visible as the bright red, chlorophyll-rich filaments binding together the cryoconite grain. Other cyanobacteria are present, including a species matching sequences from Phormidium autumnale found in an Antarctic lake. However, MinION sequencing is useful in revealing less charismatic microbes. Also abundant within the community are members of the Polaromonas genus. Found in both cold and highly polluted environments worldwide, Polaromonas bacteria are highly flexible in their lifestyles, able to adapt to using highly poisonous compounds as food sources, or even anoxygenic phototrophy (photosynthesis without using water or producing oxygen) on alpine glaciers. Cryoconite sequences matching DNA from Methylibium found in Tibetan permafrost also hint at the need for flexible metabolism to survive on glacier surfaces. Finally, Ferruginibacter sequences best matching DNA data from iron-rich dust aggregates forming on snow in the Japanese mountains suggest that cold-tolerant iron cycling may be occurring within cryoconite.
In a grain of cryoconite, we see relatives of cyanobacteria from Arctic glaciers, but also Antarctic lakes, metabolically flexible bacteria found in cold and contaminated environments, and even bacteria living by respiring iron on snow in the Japanese mountains. We see a world.
Edited by Joe Cook and Sophie Berger
 Dr Arwyn Edwards is a Senior Lecturer in Biology at Aberystwyth University and the present Royal Geographic Society’s Walters Kundert Arctic Fellow. His research on genomic diversity in glacial environments is supported by the Leverhulme Trust.
Dr Arwyn Edwards is a Senior Lecturer in Biology at Aberystwyth University and the present Royal Geographic Society’s Walters Kundert Arctic Fellow. His research on genomic diversity in glacial environments is supported by the Leverhulme Trust.