Marine picoplankton, <2 µm, are one of the most ubiquitous fauna in the open ocean. These marine microorganisms are hugely important – being responsible for a significant proportion of oceanic net primary productivity. Researchers are able to track the evolution of their genomes and the transportation of these microorganisms by analysing ice cores, which offer the potential to study the evolution of marine bacteria over approximately 300 million generations, equivalent to the last 700,000 years.
A team from Berkeley, P.B. Price and R.C. Ray, set out to map autofluorescence of chlorophyll (Chl) and the amino acid tryptophan (Trp) using fluorescence spectrometry. Autofluorescence is the ability of biological structures, such as mitochondria, to emit natural light once they have absorbed light from another source. In addition, amino acids like tryptophan show some degree of autofluorescence.
The purpose of the Berkeley study was to measure chlorophyll (Chl) and tryptophan (Trp), both of which are found in the cell walls of phytoplankton and bacteria, as a function of depth in Greenland and Antarctic ice cores. These allow scientists to measure microbial activity within a body of water and as such the biological oxygen demand. To do this they used a Scanning Fluorescence Spectrometer in the dark at -25 °C. They found that autofluorescing compounds decrease only marginally (by a factor of three) in depths in excess of ~2300 m, and that there is a clear see-saw pattern between summer and winter production, varying by about 25%.
To undertake the majority of their experiments the team used the Berkeley Fluorescence Spectrometer (BFS) to excite fluorescence in an ice core with a smoothly planed surface. In addition, they also carried out Flow Cytometric (FCM) analysis of samples cut and melted from ice cores taken from various locations on the Greenland and Antarctic ice sheets. FCM provides rapid and accurate measurements of individual phytoplanktonic cells that are too dim to be discriminated using epifluorescence microscopy, and is well suited to study the smallest size class of the plankton (<2 µm).
Their initial results reveal that cells containing Chl ecotypes (varieties specific to a particular set of environmental conditions) undergo photobleaching in air. This can complicate the observation of fluorescent molecules since they will eventually be destroyed by the light exposure that initially causes them to fluoresce, like that from a microscope beam. Furthermore, based on FCM analysis, the decline in Chl is not due to a gradual decrease of Chl autofluorescence per cell over time in the ice-bound phototrophs (organisms that carry out photon capture to acquire energy, and use the energy from light to carry out various cellular metabolic processes). It is possible then that cyanobacteria that adapt to compaction from surface snow into ice without disruption may recover from sunlight bleaching. So what was the cause?
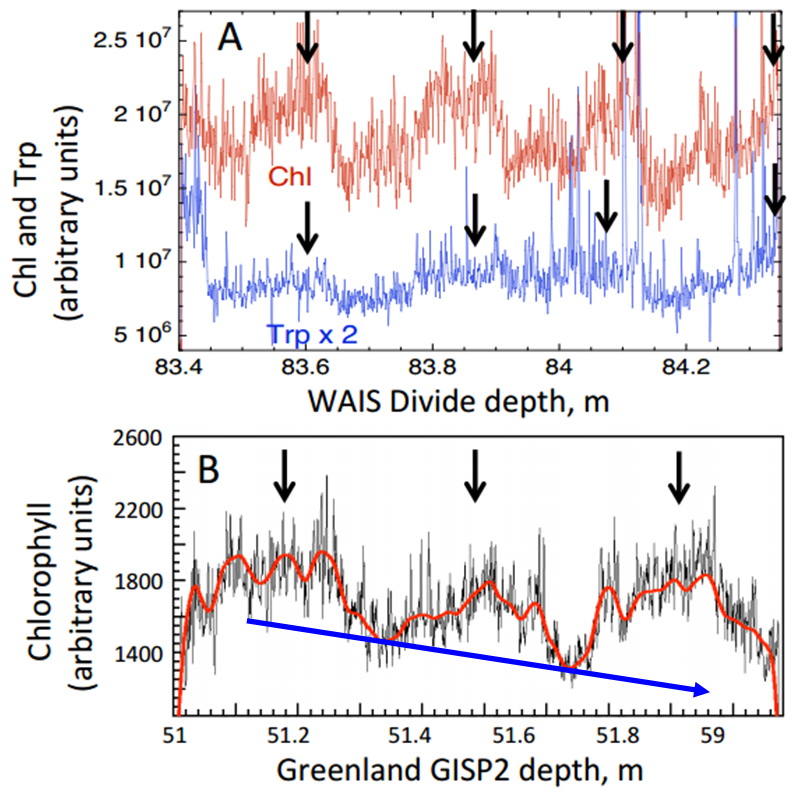
Chl autofluorescence (arbitrary units) at several depths in ice cores showing fluctuations consistent with summer-winter variations. (A) Chl and Trp over 4 yr in Western Antarctic Ice Sheet Divide ice (WAISD), summer maxima are marked with arrows; (B) Chl in Greenland ice shows maxima during 3 summers, wiggly red line is a spline fit to the data, blue arrow indicates general decrease in chlorophyll (Modified from Price and Bay 2013).
To determine whether Chl autofluorescence in the ice came from Chl in cells they used differential interference contrast microscopy and epifluorescence microscopy to search for microorganisms in melted, unstained, samples across both polar regions. Therefore, the strong Chl emission, its annual modulation in the ice cores, and the absence of Chl autofluorescence seen might be due to the presence of picocyanobacteria belonging to the genera Prochlorococcus (Pro) and Synechococcus (Syn). Buford Price comments,
“Using flow cytometry, we find that every sample of more than ~10 g of ice contains enough Pro and Syn cells that we can plot their concentration vs depth and vs location in Antarctic or Greenland ice. We can use the sorting ability of some flow cytometers to deflect only the Prochlorococcus and Synechococcus cells into sterile vials for later analysis of their DNA or for culturing”.
Syn cells are found over a much wider range than Pro cells, typically extending from pole-to-pole near nutrient-rich water, but do not extend significantly downward in the photic zone. In contrast, Pro cells have yet to be identified outside of 40°N and 40°S, and do extend down to 150 m in the photic zone. Price and Bay attribute the decrease in Chl and picocyanobacteria concentration from summer to winter to the combined effects of lower ocean temperatures and decreased daylight at latitudes populated by both Pro and Syn.
For Pro and Syn to be present at all depths in Arctic and Antarctic ice, and with a summer-winter concentration difference of only ~25%, they must have grown in waters at lower latitudes before being transported by wind and ocean currents to higher latitudes. An important discovery is that when Pro and Syn-like cells are deposited onto fresh snow and frozen into ice, FCM measurements show no obvious decrease in autofluorescence with time, with Buford Price commenting that their ultimate,
“…goal is to map changes in Pro and Syn genomes with depth and this with time in the past. This will work because at typical ice temperature at different depths (-30 to -40 C) the DNA degrades extremely slowly if the ice is reasonably pure and devoid of mineral rocks that might contain radioactive impurities” (personal communication).
This is an example of a protective feature of glacial ice that preserves Chl and other pigments in phototrophs against losing autofluorescence intensity with time in total darkness.
Their results suggest that Pro and Syn cells are transported by hemispheric wind patterns, from oceans at temperate latitudes. It is thus highly plausible that there is an annual modulation, between summer and winter. The team admit that it is very challenging to accurately model the fluxes of wind-borne Pro and Syn cells that reach the polar regions from the mid-latitudes, but the evidence that they’re transported here is convincing and presents a worthy challenge to the prevailing hypothesis that they originated separately in polar lakes.
By Alexander Stubbings, Freelance Science Writer
Reference:
Price, P. B. and Bay, R. C.: Marine bacteria in deep Arctic and Antarctic ice cores: a proxy for evolution in oceans over 300 million generations, Biogeosciences, 9, 3799-3815, 2012



