For some time now, there has been much debate about whether our beloved dinosaur, Triceratops, is a distinct species, or a younger version of a bigger ceratopsian, Torosaurus – the great Toroceratops’ debate. Proponents of both sides of the argument have made detailed quantitative and qualitative points, and there doesn’t really seem to have been any resolution. Check out the video below for a great discussion of the issues, or this link or this link.
FIGHT! (source)
Enter geometric morphometrics (GM). Everyones’ favourite-sounding method, GM is essentially a way of analysing the shape of objects, like fossils, using co-ordinate data. There are a range of statistical ways of spanking your data into shape once you have a digital version of it, and have converted an object into a series of point co-ordinates (decent-ish explanation in the methods section here).
What it is for scientists is a way of introducing statistical rigour to test a hypothesis. It’s really frickin’ useful, pretty much. The hypothesis which Maironi et al. wanted to test was whether Triceratops, Torosaurus, and Nedoceratops, another dino that might be just a younger Torosaurus, were distinct enough to not be regarded as a single growth series of the same species. Using geometric morphometrics, it is possible to visit the shape variation that specimens exhibit, as well as whether they truly represent discrete groups.
I didn’t want to this post to be critical of the methods for this paper, but they have missed a few essential steps. If you want to test for discrete clusters or groups within your data, principal components analysis (PCA) is not really the right method, as this study uses. PCA is a way of taking a high-dimensional data set, as you usually get using GM, and reducing it into fewer variables that represent a high proportion of the variance within that data set – usually, 95% of the variance is the accepted minimum required to be represented by a number of principal components.
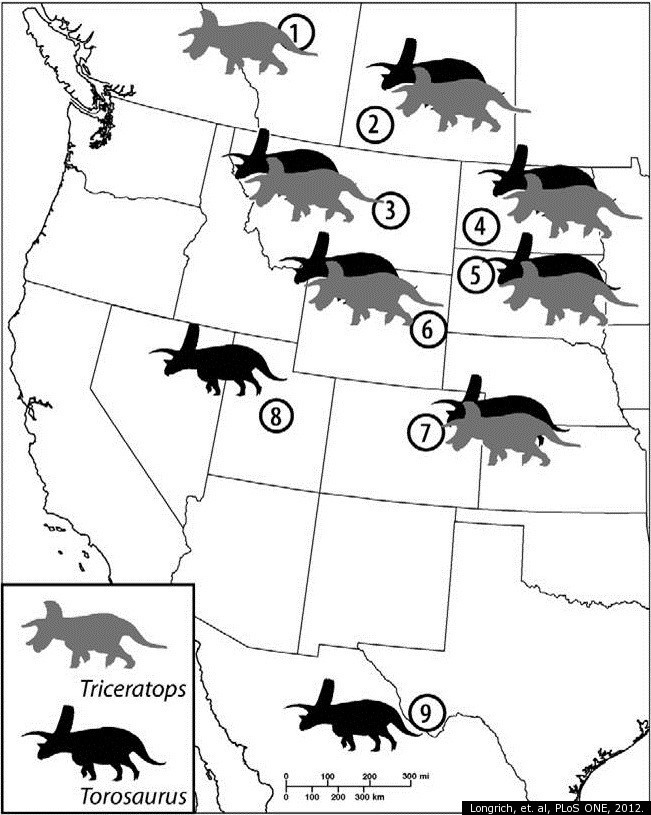
Distribution of Torosaurus and Triceratops specimens in North America (source)
In this case, 12 (!) PCs represented 95% of the variance in the reconstructed digital dinos. The first two only represent about 60% of the data – it is, then, entirely inappropriate to analyse just these two when they represent such a small proportion of the original data. What the study essentially does, is attempts to interpret distinctiveness of the different species, in a ‘morphospace’ that firstly isn’t designed to test for distinctiveness, but also fails to faithfully represent the data. Ideally, you’d want to analyse the dispersion pattern of all of the specimens with respect to the first 12 axes – this would tell you about the shape changes that are being represented by each axis.
The next step should have been to use a form off discriminant analysis – these tests are designed to test for group distinctiveness, as they incorporate group-level information as an additional variable; in this case, the species’ ID. Performing this would tell you how accurate, or distinct, your supposedly different groups are, and you can test the proportion of specimens that are correctly ‘assigned’ to each group. I go into this protocol in some detail in a forthcoming paper, and will discuss it more then. For now, I feel a bit bad saying, but this is really only half an analysis, and a lot more could have been and should have been done with it.
Nonetheless, there are some interesting results, if not based on an entirely substantial analysis (imo). Importantly, Torosaurus and Triceratops, in the context of the first two principal components, occupy different morphospaces, which suggests that they can be delimited based on shape as well as their sizes. That’s pretty much it. Another piece of the puzzle suggesting that we can keep Triceratops alive, for now!
Fortunately, the authors have been jolly decent uploaded their raw data, so I’m gonna go have a play with it and see what we get.. BRB
Reference:
Maiorini, L., Farke, A. A., Kotsakis, T. and Piras, P. (2013) Is Torosaurus Triceratops? Geometric morphometric evidence of Late Maastrichtian ceratopsid dinosaurs, PLOS ONE, 8(11), e81608 (OA link)


David Button
Good post. I’ll be interested to see how your ultimate findings differ.
Mike Keesey
“Another piece of the puzzle suggesting that we can keep Triceratops alive, for now!”
I’m sure you know this, but it’s Torosaurus Marsh 1891 that dies if they’re synonymized, not Triceratops Marsh 1889.
Jon Tennant
Oh, yeah, of course.
I actually really don’t like this, as it either means we have to diagnose Triceratops based on immature material, or transfer the diagnosis for Torosaurus over. I think we should just dissolve Triceratops 😉
Mike Keesey
Tremendously unstable idea. You know how many taxa are based on less-than-mature holotypes? Off the top of my head, Australopithecus, Homo habilis, Homo ergaster, Compsognathus, Epidendrosaurus, Scipionyx, Mussaurus….
In some of these cases you could possibly make the argument for a neotype. Although, come on, even if the Triceratops type material is developmentally subadult, it’s still mostly grown and probably sexually mature. How mature is mature enough?
Nick
I’m certain you meant to write “suggesting we can keep Torosaurus alive for now”.
Nick
And ugh, no comments were loaded for me at the time, so I never saw Keesey’s comment.
Pingback: Is Torosaurus Triceratops? The debate rages on!...
Antoine Souron
Thank you for this interesting post!
I had a quick look at the article, and IMO, there are a bigger problems than the number of PC considered or the absence of discriminant analyses.
If I understood correctly, they used 2D landmarks and semi-landmarks digitized on photographs to compare the different specimens. In my research on Suidae, I used 3D landmarks to quantify skull shape, and I compared it to previous work that used 2D landmarks digitized on photographs. And unsurprisingly, there were many differences in the results. Trying to characterize a 3D object using 2D photographs seems very difficult and subject to various biases (notably differences in orientation, in perspectives…).
Otherwise, that sounds like an interesting work (I look forward to reading the next articles by the Berkeley team on the ontogeny).
Jon Tennant
Hey Antione, thanks for your comment.
I believe the authors do address why they think 2D is an appropriate method in their paper – I can’t recall the reasons off the top of my head though.
There was a cool article out very recently by Francois Gould investigating the functional implications of 2D vs 3D morphometrics – worth checking out! http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0091719
Antoine Souron
Thanks for the very useful reference (I had missed it!). Gould found that both linear measurements and GM surface analysis enabled to distinguish between the femora of mammals displaying different modes of locomotion (GM was a slightly more discriminating). I think he is trying to draw attention to the need of balancing the time and cost of analyses against the power of discrimation (I guess the linear measurements win in that case).
Still, I think GM is way superior in terms of visualization tools and the possibility of analyzing differences in shape and size independently. Again, that’s my opinion based on my own research on skulls of Suidae, and I am sure it depends on which group and which body parts you study.
Pingback: Morsels For The Mind – 21/03/2014 › Six Incredible Things Before Breakfast